Nef HIV-1 (Retroviridae: Orthoretrovirinae: Lentivirus: Human immunodeficiency virus-1), multifunctional protein: features of genetic virus variants circulating in Russia
- Authors: Kuznetsova A.I.1, Antonova A.A.1, Protasova L.A.1, Glyakina A.V.2, Kim K.V.1, Munchak I.M.1, Mezhenskaya E.N.1, Orlova-Morozova E.A.3, Pronin A.Y.3, Prilipov A.G.1, Galzitskaya O.V.1,4
-
Affiliations:
- D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
- Institute of Mathematical Problems of Biology RAS – the Branch of Keldysh Institute of Applied Mathematics of Russian Academy of Sciences
- Center for the Prevention and Control of AIDS and Infectious Diseases
- Institute of Theoretical and Experimental Biophysics RAS
- Issue: Vol 70, No 6 (2025)
- Pages: 518-535
- Section: ORIGINAL RESEARCHES
- URL: https://ogarev-online.ru/0507-4088/article/view/375502
- DOI: https://doi.org/10.36233/0507-4088-305
- EDN: https://elibrary.ru/udlkyb
- ID: 375502
Cite item
Abstract
Introduction. Nef provides high level of HIV-1 replication due to synergy of its multiple functions and is an important factor in the pathogenesis of HIV infection. Nef is considered as a target for development of therapeutic agents. Mutations of drug resistance to dolutegravir can occur in Nef protein. Natural amino acid substitutions in Nef protein have been associated with the degree of progression of HIV infection, development of neurodegenerative and cardiovascular diseases in patients.
The aim of the study is to investigate Nef genetic diversity in HIV-1 variants circulating in Russia and in Moscow region.
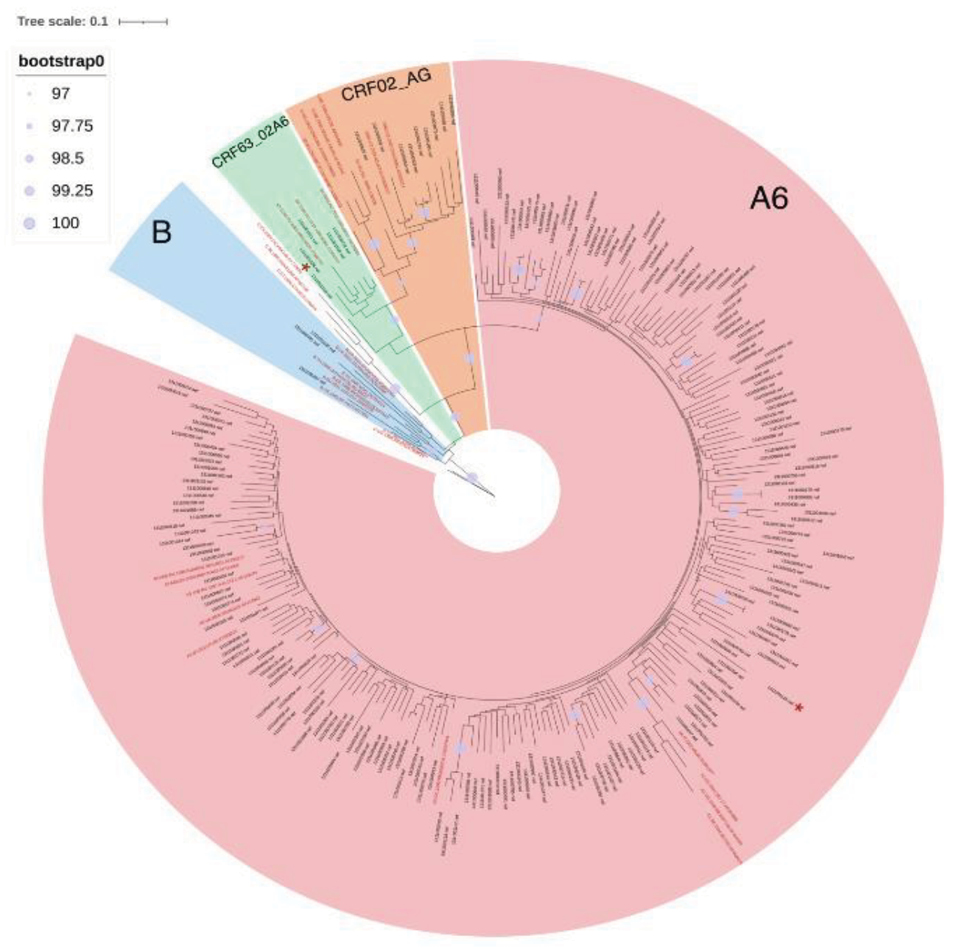
Materials and methods. Total 216 Nef sequences obtained from whole blood samples of patients and 77 sequences downloaded from the Los Alamos International Database were analyzed. Consensus sequences of Nef sub-subtype A6, subtype B, CRF02_AG, CRF63_02A6, CRF133_A6B, and the reference sequence NL4-3 were compared. Genetic diversity of Nef sub-subtype A6 (Nef-A6) in patients with different stages of the disease was assessed. The presence of dolutegravir-associated drug resistance mutations in the Nef protein in HIV-1 variants circulating in Russia was also investigated.
Results. Differences in the spatial structures in consensus sequences of the studied HIV-1 variants were determined. It was shown that the conservatism of Nef-A6 in groups of patients with later stages of the disease was significantly higher. No mutations of drug resistance to dolutegravir were detected.
Conclusion. The differences in Nef sequences of HIV-1 variants circulating in Russia could affect the functional properties of the protein and could be taken into account in creating Nef-based therapies in the future. Obtained results indicate that there is no risk of resistance to dolutegravir associated with – mutations in the Nef protein. It outlines possible directions for further research into the genetic diversity of Nef.
Keywords
About the authors
Anna I. Kuznetsova
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Email: a-myznikova@list.ru
ORCID iD: 0000-0001-5299-3081
head of laboratory of T-lymphotropic viruses, PhD, leading researcher
Russian Federation, 123098, MoscowAnastasiia A. Antonova
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Email: anastaseika95@mail.ru
ORCID iD: 0000-0002-9180-9846
PhD, Researcher, Laboratory of T-lymphotropic viruses
Russian Federation, 123098, MoscowLarisa A. Protasova
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Email: larisa.protasova.03@mail.ru
ORCID iD: 0009-0001-0430-1578
research assistant, Laboratory of T-lymphotropic viruses
Russian Federation, 123098, MoscowAnna V. Glyakina
Institute of Mathematical Problems of Biology RAS – the Branch of Keldysh Institute of Applied Mathematics of Russian Academy of Sciences
Email: quark777a@gmail.com
ORCID iD: 0000-0002-6352-2880
PhD, Researcher
Russian Federation, 142290, PushchinoKristina V. Kim
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Email: kimsya99@gmail.com
ORCID iD: 0000-0002-4150-2280
junior researcher, Laboratory of T-lymphotropic viruses
Russian Federation, 123098, MoscowIana M. Munchak
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Email: yana_munchak@mail.ru
ORCID iD: 0000-0002-4792-8928
junior researcher, Laboratory of T-lymphotropic viruses
Russian Federation, 123098, MoscowEkaterina N. Mezhenskaya
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Email: belokopytova.01@mail.ru
ORCID iD: 0000-0002-3110-0843
PhD, Researcher, Laboratory of T-lymphotropic viruses
Russian Federation, 123098, MoscowElena A. Orlova-Morozova
Center for the Prevention and Control of AIDS and Infectious Diseases
Email: orlovamorozova@gmail.com
ORCID iD: 0000-0003-2495-6501
PhD, Head of outpatient department
Russian Federation, 140053, KotelnikiAlexander Yu. Pronin
Center for the Prevention and Control of AIDS and Infectious Diseases
Email: alexanderp909@gmail.com
ORCID iD: 0000-0001-9268-4929
PhD, Chief Physician
Russian Federation, 140053, KotelnikiAlexey G. Prilipov
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Email: a_prilipov@mail.ru
ORCID iD: 0000-0001-8755-1419
Doctor of Biological Sciences, leading researcher, head of the laboratory of molecular genetics
Russian Federation, 123098, MoscowOxana V. Galzitskaya
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya; Institute of Theoretical and Experimental Biophysics RAS
Author for correspondence.
Email: ogalzit@vega.protres.ru
ORCID iD: 0000-0002-3962-1520
Doctor of Physical and Mathematical Sciences, Head of the Bioinformatics Laboratory, Chief Researcher
Russian Federation, 123098, Moscow; 142290, PushchinoReferences
- Pereira E.A., da Silva L.L. HIV-1 Nef: taking control of protein trafficking. Traffic. 2016;17(9): 976–96. https://doi.org/10.1111/tra.12412
- Raymond A.D., Campbell-Sims T.C., Khan M., Lang M., Huang M.B., Bond V.C., et al. HIV Type 1 Nef is released from infected cells in CD45(+) microvesicles and is present in the plasma of HIV-infected individuals. AIDS Res. Hum. Retroviruses. 2011; 27(2): 167–78. https://doi.org/10.1089/aid.2009.0170
- Udenwobele D.I., Su R.C., Good S.V., Ball T.B., Varma Shrivastav S., Shrivastav A. Myristoylation: an important protein modification in the immune response. Front. Immunol. 2017; 8: 751. https://doi.org/10.3389/fimmu.2017.00751
- Foster J.L., Denial S.J., Temple B.R., Garcia J.V. Mechanisms of HIV-1 Nef function and intracellular signaling. J. Neuroimmune Pharmacol. 2011; 6(2): 230–46. https://doi.org/10.1007/s11481-011-9262-y
- Arold S., Franken P., Strub M.P., Hoh F., Benichou S., Benarous R., et al. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure. 1997; 5(10): 1361–72. https://doi.org/10.1016/s0969-2126(97)00286-4
- Lee C.H., Saksela K., Mirza U.A., Chait B.T., Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996; 85(6): 931–42. https://doi.org/10.1016/s0092-8674(00)81276-3
- Grzesiek S., Bax A., Hu J.S., Kaufman J., Palmer I., Stahl S.J., et al. Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Sci. 1997; 6(6): 1248–63. https://doi.org/10.1002/pro.5560060613
- Grzesiek S., Stahl S.J., Wingfield P.T., Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996; 35(32): 10256–61. https://doi.org/10.1021/bi9611164
- Geyer M., Munte C.E., Schorr J., Kellner R., Kalbitzer H.R. Structure of the anchor-domain of myristoylated and non-myristoylated HIV-1 Nef protein. J. Mol. Biol. 1999; 289(1): 123–38. https://doi.org/10.1006/jmbi.1999.2740
- Kirchhoff F., Schindler M., Bailer N., Renkema G.H., Saksela K., Knoop V., et al. Nef proteins from simian immunodeficiency virus-infected chimpanzees interact with p21-activated kinase 2 and modulate cell surface expression of various human receptors. J. Virol. 2004; 78(13): 6864–74. https://doi.org/10.1128/JVI.78.13.6864-6874.2004
- Corró G., Rocco C.A., De Candia C., Catano G., Turk G., Mangano A., et al. Genetic and functional analysis of HIV type 1 Nef gene derived from long-term nonprogressor children: association of attenuated variants with slow progression to pediatric AIDS. AIDS Res. Hum. Retroviruses. 2012; 28(12): 1617–26. https://doi.org/10.1089/AID.2012.0020
- Chen Y.L., Trono D., Camaur D. The proteolytic cleavage of human immunodeficiency virus type 1 Nef does not correlate with its ability to stimulate virion infectivity. J. Virol. 1998; 72(4): 3178–84. https://doi.org/10.1128/JVI.72.4.3178-3184.1998
- Toyoda M., Ogata Y., Mahiti M., Maeda Y., Kuang X.T., Miura T., et al. Differential ability of primary HIV-1 Nef isolates to downregulate HIV-1 entry receptors. J. Virol. 2015; 89(18): 9639–52. https://doi.org/10.1128/JVI.01548-15
- Saksela K., Cheng G., Baltimore D. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 1995; 14(3): 484–91. https://doi.org/10.1002/j.1460-2075.1995.tb07024.x
- Malet I., Subra F., Charpentier C., Collin G., Descamps D., Calvez V., et al. Mutations located outside the integrase gene can confer resistance to HIV-1 integrase strand transfer inhibitors. mBio. 2017; 8(5): e00922-17. https://doi.org/10.1128/mBio.00922-17
- Poe J.A., Smithgall T.E. HIV-1 Nef dimerization is required for Nef-mediated receptor downregulation and viral replication. J. Mol. Biol. 2009; 394(2): 329–42. https://doi.org/10.1016/j.jmb.2009.09.047
- Liu L.X., Heveker N., Fackler O.T., Arold S., Le Gall S., Janvier K., et al. Mutation of a conserved residue (D123) required for oligomerization of human immunodeficiency virus type 1 Nef protein abolishes interaction with human thioesterase and results in impairment of Nef biological functions. J. Virol. 2000; 74(11): 5310–9. https://doi.org/10.1128/jvi.74.11.5310-5319.2000
- Stolp B., Fackler O.T. How HIV takes advantage of the cytoskeleton in entry and replication. Viruses. 2011; 3(4): 293–311. https://doi.org/10.3390/v3040293
- Schindler M., Rajan D., Specht A., Ritter C., Pulkkinen K., Saksela K., et al. Association of Nef with p21-activated kinase 2 is dispensable for efficient human immunodeficiency virus type 1 replication and cytopathicity in ex vivo-infected human lymphoid tissue. J. Virol. 2007; 81(23): 13005–14. https://doi.org/10.1128/JVI.01436-07
- Lama J., Ware C.F. Human immunodeficiency virus type 1 Nef mediates sustained membrane expression of tumor necrosis factor and the related cytokine LIGHT on activated T cells. J. Virol. 2000; 74(20): 9396–402. https://doi.org/10.1128/jvi.74.20.9396-9402.2000
- Smith D.M., Richman D.D., Little S.J. HIV superinfection. J. Infect. Dis. 2005; 192(3): 438–44. https://doi.org/10.1086/431682
- Michel N., Allespach I., Venzke S., Fackler O.T., Keppler O.T. The Nef protein of human immunodeficiency virus establishes superinfection immunity by a dual strategy to downregulate cell-surface CCR5 and CD4. Curr. Biol. 2005; 15(8): 714–23. https://doi.org/10.1016/j.cub.2005.02.058
- Schwartz O., Maréchal V., Le Gall S., Lemonnier F., Heard J.M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 1996; 2(3): 338–42. https://doi.org/10.1038/nm0396-338
- Stumptner-Cuvelette P., Morchoisne S., Dugast M., Le Gall S., Raposo G., Schwartz O., et al. HIV-1 Nef impairs MHC class II antigen presentation and surface expression. Proc. Natl. Acad. Sci. USA. 2001; 98(21): 12144–9. https://doi.org/10.1073/pnas.221256498
- Stove V., Van de Walle I., Naessens E., Coene E., Stove C., Plum J., et al. Human immunodeficiency virus Nef induces rapid internalization of the T-cell coreceptor CD8alphabeta. J. Virol. 2005; 79(17): 11422–33. https://doi.org/10.1128/JVI.79.17.11422-11433.2005
- Cole D.K., Laugel B., Clement M., Price D.A., Wooldridge L., Sewell A.K. The molecular determinants of CD8 co-receptor function. Immunology. 2012; 137(2): 139–48. https://doi.org/10.1111/j.1365-2567.2012.03625.x
- Swigut T., Shohdy N., Skowronski J. Mechanism for down-regulation of CD28 by Nef. EMBO J. 2001; 20(7): 1593–604. https://doi.org/10.1093/emboj/20.7.1593
- Usami Y., Wu Y., Göttlinger H.G. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015; 526(7572): 218–23. https://doi.org/10.1038/nature15400
- Rosa A., Chande A., Ziglio S., De Sanctis V., Bertorelli R., Goh S.L., et al. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature. 2015; 526(7572): 212–7. https://doi.org/10.1038/nature15399
- Popova N.V., Deyev I.E., Petrenko A.G. Clathrin-mediated endocytosis and adaptor proteins. Acta Naturae. 2013; 5(3): 62–73. https://doi.org/10.32607/20758251-2013-5-3-62-73 https://elibrary.ru/rvzywf
- Chou I.J., Hou J.Y., Fan W.L., Tsai M.H., Lin K.L. Long-term outcome of neonatal seizure with PACS2 mutation: case series and literature review. Children (Basel). 2023; 10(4): 621. https://doi.org/10.3390/children10040621
- Roskoski R. Jr. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol Res. 2015; 94: 9–25. https://doi.org/10.1016/j.phrs.2015.01.003
- Francis C.R., Bell M.L., Skripnichuk M.M., Kushner E.J. Arf6 is required for endocytosis and filamentous actin assembly during angiogenesis in vitro. Microcirculation. 2023; 30(8): e12831. https://doi.org/10.1111/micc.12831
- Wan L., Molloy S.S., Thomas L., Liu G., Xiang Y., Rybak S.L., et al. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998; 94(2): 205–16. https://doi.org/10.1016/s0092-8674(00)81420-8
- Usmani S.M., Murooka T.T., Deruaz M., Koh W.H., Sharaf R.R., Di Pilato M., et al. HIV-1 balances the fitness costs and benefits of disrupting the host cell actin cytoskeleton early after mucosal transmission. Cell Host Microbe. 2019; 25(1): 73–86.e5. https://doi.org/10.1016/j.chom.2018.12.008
- Ostrowska Z., Moraczewska J. Cofilin – a protein controlling dynamics of actin filaments. Postepy Hig. Med. Dosw. (Online). 2017; 71(0): 339–51. https://doi.org/10.5604/01.3001.0010.3818
- Aqil M., Naqvi A.R., Bano A.S., Jameel S. The HIV-1 Nef protein binds argonaute-2 and functions as a viral suppressor of RNA interference. PLoS One. 2013; 8(9): e74472. https://doi.org/10.1371/journal.pone.0074472
- Qiao X., He B., Chiu A., Knowles D.M., Chadburn A., Cerutti A. Human immunodeficiency virus 1 Nef suppresses CD40-dependent immunoglobulin class switching in bystander B cells. Nat. Immunol. 2006; 7(3): 302–10. https://doi.org/10.1038/ni1302
- Olivetta E., Arenaccio C., Manfredi F., Anticoli S., Federico M. The contribution of extracellular Nef to HIV-induced pathogenesis. Curr. Drug Targets. 2016; 17(1): 46–53. https://doi.org/10.2174/1389450116666151001110126
- Sami Saribas A., Cicalese S., Ahooyi T.M., Khalili K., Amini S., Sariyer I.K. HIV-1 Nef is released in extracellular vesicles derived from astrocytes: evidence for Nef-mediated neurotoxicity. Cell Death Dis. 2017; 8(1): e2542. https://doi.org/10.1038/cddis.2016.467
- Yarandi S.S., Duggan M.R., Sariyer I.K. Emerging role of Nef in the development of HIV associated neurological disorders. J. Neuroimmune Pharmacol. 2021; 16(2): 238–50. https://doi.org/10.1007/s11481-020-09964-1
- Cruz N.V., Amorim R., Oliveira F.E., Speranza F.A., Costa L.J. Mutations in the Nef and Vif genes associated with progression to AIDS in elite controller and slow-progressor patients. J. Med. Virol. 2013; 85(4): 563–74. https://doi.org/10.1002/jmv.23512
- Kirchhoff F., Easterbrook P.J., Douglas N., Troop M., Greenough T.C., Weber J., et al. Sequence variations in human immunodeficiency virus type 1 Nef are associated with different stages of disease. J. Virol. 1999; 73(7): 5497–508. https://doi.org/10.1128/JVI.73.7.5497-5508.1999
- Lamers S.L., Poon A.F., McGrath M.S. HIV-1 Nef protein structures associated with brain infection and dementia pathogenesis. PLoS One. 2011; 6(2): e16659. https://doi.org/10.1371/journal.pone.0016659
- Almodovar S., Knight R., Allshouse A.A., Roemer S., Lozupone C., McDonald D., et al. Human Immunodeficiency Virus Nef signature sequences are associated with pulmonary hypertension. AIDS Res. Hum. Retroviruses. 2012; 28(6): 607–18. https://doi.org/10.1089/AID.2011.0021
- Emert-Sedlak L.A., Loughran H.M., Shi H., Kulp J.L. 3rd, Shu S.T., Zhao J., et al. Synthesis and evaluation of orally active small molecule HIV-1 Nef antagonists. Bioorg. Med. Chem. Lett. 2016; 26(5): 1480–4. https://doi.org/10.1016/j.bmcl.2016.01.043
- Hunegnaw R., Vassylyeva M., Dubrovsky L., Pushkarsky T., Sviridov D., Anashkina A.A., et al. Interaction between HIV-1 Nef and Calnexin: from modeling to small molecule inhibitors reversing HIV-induced lipid accumulation. Arterioscler. Thromb. Vasc. Biol. 2016; 36(9): 1758–71. https://doi.org/10.1161/ATVBAHA.116.307997
- Milani A., Baesi K., Agi E., Marouf G., Ahmadi M., Bolhassani A. HIV-1 accessory proteins: which one is potentially effective in diagnosis and vaccine development? Protein Pept. Lett. 2021; 28(6): 687–98. https://doi.org/10.2174/0929866528999201231213610
- Kardani K., Hashemi A., Bolhassani A. Comparison of HIV-1 Vif and Vpu accessory proteins for delivery of polyepitope constructs harboring Nef, Gp160 and P24 using various cell penetrating peptides. PLoS One. 2019; 14(10): e0223844. https://doi.org/10.1371/journal.pone.0223844
- Lebedev A., Kireev D., Kirichenko A., Mezhenskaya E., Antonova A., Bobkov V., et al. The molecular epidemiology of HIV-1 in Russia, 1987–2023: subtypes, transmission networks and phylogenetic story. Pathogens. 2025; 14(8): 738. https://doi.org/10.3390/pathogens14080738
- Halikov M.R., Ekushov V.E., Totmenin A.V., Gashnikova N.M., Antonets M.E., Tregubchak T.V., et al. Identification of a novel HIV-1 circulating recombinant form CRF157_A6C in Primorsky Territory, Russia. J. Infect. 2024; 88(2): 180–2. https://doi.org/10.1016/j.jinf.2023.11.005
- Antonova A.A., Kuznetsova A.I., Ozhmegova E.N., Lebedev A.V., Kazennova E.V., Kim K.V., et al. Genetic diversity of HIV-1 at the current stage of the epidemic in the Russian Federation: an increase in the prevalence of recombinant forms. VICH-infektsiya i immunosupressii. 2023; 15(3): 61–72. https://doi.org/10.22328/2077-9828-2023-15-3-61-72 https://elibrary.ru/tpwttn (in Russian)
- Maksimenko L.V., Sivay M.V., Totmenin A.V., Shvalov A.N., Skudarnov S.E., Ostapova T.S., et al. Novel HIV-1 A6/B recombinant forms (CRF133_A6B and URF_A6/B) circulating in Krasnoyarsk region, Russia. J. Infect. 2022; 85(6): 702–69. https://doi.org/10.1016/j.jinf.2022.10.001
- Gromov K.B., Kireev D.E., Murzakova A.V., Lopatukhin A.E., Kazennova E.V., Bobkova M.R. Analysis of HIV-1 (Human immunodeficiency virus-1, Lentivirus, Orthoretrovirinae, Retroviridae) Nef protein polymorphism of variants circulating in the former USSR countries. Voprosy virusologii. 2019; 64(6): 281–90. https://doi.org/10.36233/0507-4088-2019-64-6-281-290 https://elibrary.ru/ukpfsl (in Russian)
- Antonova A.A., Lebedev A.V., Ozhmegova E.N., Shlykova A.V., Lapavok I.A., Kuznetsova A.I. Variability of non-structural proteins in HIV-1 sub-subtype A6 (Retroviridae: Orthoretrovirinae: Lentivirus: Human immunodeficiency virus-1, sub-subtype A6) variants circulating in different regions of the Russian Federation. Voprosy virusologii. 2024; 69(5): 470–80. https://doi.org/10.36233/0507-4088-262 https://elibrary.ru/wbbkuq (in Russian)
- Ryzhov K.A., Nosik M.N., Kravtchenko A.V. Study of the HIV-1 regulatory genes using the polymerase chain reaction. Voprosy virusologii. 2015; 60(3): 41–4. https://elibrary.ru/tsztej (in Russian)
- HIV infection in adults. Clinical guidelines; 2024. Available at: https://cr.minzdrav.gov.ru/preview-cr/79_2?ysclid=m7vwu0c9ot784280780 (in Russian)
- Antonova A.A., Protasova L.A., Kim K.V., Munchak Ia.M., Mezhenskaya E.N., Orlova-Morozova E.A., et al. Genetic diversity of Vif protein in human immunodeficiency virus type 1 variants (Retroviridae: Orthoretrovirinae: Lentivirus: Human immunodeficiency virus-1) that circulated in the Moscow region in 2019–2020. Voprosy virusologii. 2025; 70(2): 117–32. https://doi.org/10.36233/0507-4088-281 https://elibrary.ru/qoqqce (in Russian)
- Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014; 30(22): 3276–8. https://doi.org/10.1093/bioinformatics/btu531
- Lobanov M.Y., Sokolovskiy I.V., Galzitskaya O.V. IsUnstruct: prediction of the residue status to be ordered or disordered in the protein chain by a method based on the Ising model. J. Biomol. Struct. Dyn. 2013; 31(10): 1034–43. https://doi.org/10.1080/07391102.2012.718529
- Lobanov M.Y., Pereyaslavets L.B., Likhachev I.V., Matkarimov B.T., Galzitskaya O.V. Is there an advantageous arrangement of aromatic residues in proteins? Statistical analysis of aromatic interactions in globular proteins. Comput. Struct. Biotechnol. J. 2021; 19: 5960–8. https://doi.org/10.1016/j.csbj.2021.10.036
- Berezov T.T., Korovkin B.F. Biological Chemistry [Biologicheskaya khimiya]. Moscow: Meditsina; 1998. https://elibrary.ru/zrnxkv (in Russian)
Supplementary files