CRISPR-Cas genome editing system in the diagnosis and therapy of infection caused by herpes simplex virus type 1 (Orthoherpesviridae: Alphaherpesviridae: Simplexvirus: Simplexvirus humanalpha1)
- Authors: Demidova N.A.1, Klimova R.R.1, Kushch A.A.1, Karpov D.S.2
-
Affiliations:
- Gamaleya National Research Centre for Epidemiology and Microbiology, Ministry of Health of the Russian Federation
- Engelhardt Institute of Molecular Biology, Russian Academy of Sciences
- Issue: Vol 70, No 6 (2025)
- Pages: 493-507
- Section: REVIEWS
- URL: https://ogarev-online.ru/0507-4088/article/view/375500
- DOI: https://doi.org/10.36233/0507-4088-307
- EDN: https://elibrary.ru/udjxmq
- ID: 375500
Cite item
Abstract
Herpes simplex virus type 1 (HSV-1), newly named as Simplexvirus humanalpha1 is one of the most common pathogens in the human population, which can cause severe disease, often with fatal outcomes. Diagnostic methods currently in use are specific and sensitive, but time-consuming, require expensive laboratory equipment and highly qualified personnel. Existing therapeutic agents have a number of significant drawbacks. To successfully treat and prevent the spread of the infection, new rapid, easy-to-use, and highly sensitive diagnostic tools and effective therapeutic agents are required. One approach to achieve this goal is CRISPR-based technology.
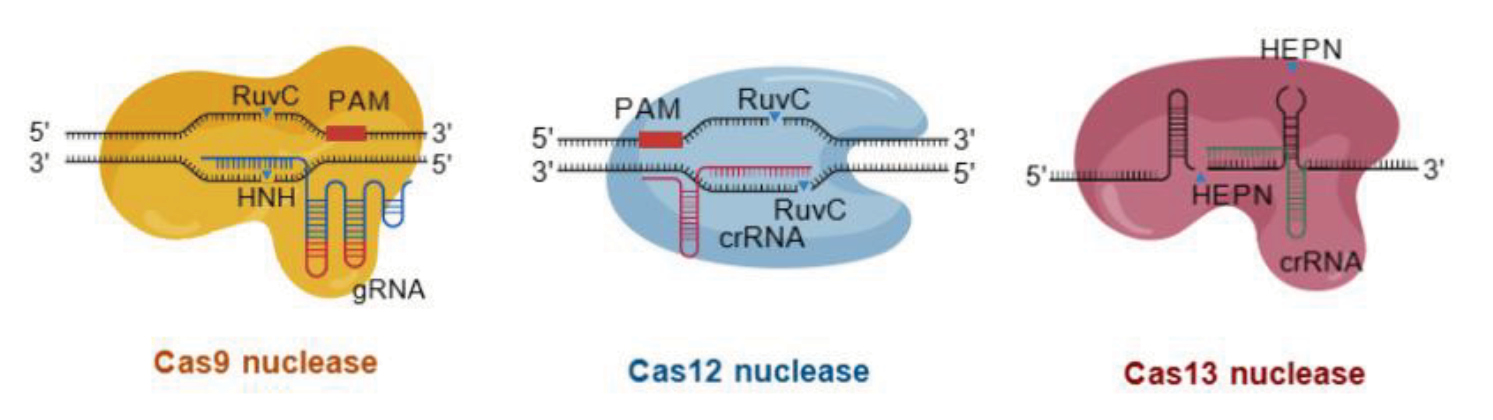
This review analyzes information obtained from a literature search in the Scopus, Web of Science and MedLine databases on the topics «HSV-1, structure, distribution, life cycle», «new methods for molecular diagnosis of HSV-1-infection», «classification of CRISPR-Cas systems», «nucleic acid amplification methods», «CRISPR-Cas effector proteins», «application of CRISPR-Cas systems in molecular diagnostics of HSV-1-infection», «application of CRISPR-Cas systems in therapy of HSV-1-infection». New approaches of CRISPR using effector proteins Cas12 and Cas13 in the diagnosis of HSV-1 infections are reviewed. The article discusses the progress in the development of CRISPR-Cas-based therapies against HSV-1-infection in vitro and in vivo. CRISPR gene therapy in vivo has a great clinical potential, but its safety and efficacy require further investigation. An analysis of the available data suggests that CRISPR-based technologies offer promising prospects for expanding the arsenal of diagnostic tools and antiviral drugs in the context of current and future outbreaks of viral diseases.
About the authors
Natalia A. Demidova
Gamaleya National Research Centre for Epidemiology and Microbiology, Ministry of Health of the Russian Federation
Email: ailande@yandex.ru
ORCID iD: 0000-0003-1961-9789
Researcher of the Laboratory of Cell Engineering
Russian Federation, 123098, MoscowRegina R. Klimova
Gamaleya National Research Centre for Epidemiology and Microbiology, Ministry of Health of the Russian Federation
Email: regi.K@mail.ru
ORCID iD: 0000-0002-4147-8444
PhD (Biology), Senior Researcher of the Laboratory of Cell Engineering
Russian Federation, 123098, MoscowAlla A. Kushch
Gamaleya National Research Centre for Epidemiology and Microbiology, Ministry of Health of the Russian Federation
Email: vitallku@mail.ru
ORCID iD: 0000-0002-3396-5533
D.Sci. (Biology), Professor, Chief Researcher of the Laboratory of Cell Engineering
Russian Federation, 123098, MoscowDmitry S. Karpov
Engelhardt Institute of Molecular Biology, Russian Academy of Sciences
Author for correspondence.
Email: aleom@yandex.ru
ORCID iD: 0000-0001-5203-0787
PhD (Biology), Leading Researcher
Russian Federation, 119991, MoscowReferences
- James C., Harfouche M., Welton N.J., Turner K.M., Abu-Raddad L.J., Gottlieb S.L., et al. Herpes simplex virus: global infection prevalence and incidence estimates, 2016. Bull. World Health Organ. 2020; 98(5): 315. https://doi.org/10.2471/BLT.19.237149
- Marcocci M.E., Napoletani G., Protto V., Kolesova O., Piacentini R., Li Puma D.D., et al. Herpes simplex virus-1 in the brain: the dark side of a sneaky infection. Trends Microbiol. 2020; 28(10): 808. https://doi.org/10.1016/j.tim.2020.03.003
- de Sousa R.M.P., Garcia L.S., Lemos F.S., de Campos V.S., Machado Ferreira E., de Almeida N.A.A., et al. CRISPR/Cas9 eye drop HSV-1 treatment reduces brain viral load: a novel application to prevent neuronal damage. Pathogens. 2024; 13(12): 1087. https://doi.org/10.3390/pathogens13121087
- Stahl J.P., Mailles A. Herpes simplex virus encephalitis update. Curr. Opin. Infect. Dis. 2019; 32(3): 239. https://doi.org/10.1097/QCO.0000000000000554
- Akkaya O. Prevalence of herpes simplex virus infections in the central nervous system. Clin. Lab. 2021; 67(7). https://doi.org/10.7754/Clin.Lab.2020.201111
- Rippee-Brooks M.D., Wu W., Dong J., Pappolla M., Fang X., Bao X. Viral infections, are they a trigger and risk factor of Alzheimer’s disease? Pathogens. 2024; 13(3): 240. https://doi.org/10.3390/pathogens13030240
- Vestin E., Bostrom G., Olsson J., Elgh F., Lind L., Kilander L., et al. Herpes simplex viral infection doubles the risk of dementia in a contemporary cohort of older adults: a prospective study. J. Alzheimers Dis. 2024; 97(4): 1841. https://doi.org/10.3233/JAD-230718
- Van Wagoner N., Qushair F., Johnston C. Genital herpes infection: progress and problems. Infect. Dis. Clin. North Am. 2023; 37(2): 351. https://doi.org/10.1016/j.idc.2023.02.011
- Wang H. Practical updates in clinical antiviral resistance testing. J. Clin. Microbiol. 2024; 62(8): e0072823. https://doi.org/10.1128/jcm.00728-23
- Sadowski L.A., Upadhyay R., Greeley Z.W., Margulies B.J. Current drugs to treat infections with herpes simplex viruses-1 and -2. Viruses. 2021; 13(7): 1228. https://doi.org/10.3390/v13071228
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337(6096): 816. https://doi.org/10.1126/science.1225829
- van Diemen F.R., Kruse E.M., Hooykaas M.J., Bruggeling C.E., Schurch A.C., van Ham P.M., et al. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016; 12(6): e1005701. https://doi.org/10.1371/journal.ppat.1005701
- Zhang I., Hsiao Z., Liu F. Development of genome editing approaches against herpes simplex virus infections. Viruses. 2021; 13(2): 338. https://doi.org/10.3390/v13020338
- Karpov D.S., Demidova N.A., Kulagin K.A., Shuvalova A.I., Kovalev M.A., Simonov R.A., et al. Complete and prolonged inhibition of herpes simplex virus type 1 infection in vitro by CRISPR/Cas9 and CRISPR/CasX systems. Int. J. Mol. Sci. 2022; 23(23): 14847. https://doi.org/10.3390/ijms232314847
- Dou B., Zhang Y., Gao H., Zhang S., Zheng J., Lu X., et al. CRISPR/Cas12a-based MUSCA-PEC strategy for HSV-1 assay. Anal. Chim. Acta. 2023; 1250: 340955. https://doi.org/10.1016/j.aca.2023.340955
- Du K., Zeng Q., Jiang M., Hu Z., Zhou M., Xia K. CRISPR/Cas12a-based biosensing: advances in mechanisms and applications for nucleic acid detection. Biosensors (Basel). 2025; 15(6): 360. https://doi.org/10.3390/bios15060360
- Packard J.E., Dembowski J.A. HSV-1 DNA replication-coordinated regulation by viral and cellular factors. Viruses. 2021; 13(10): 2015. https://doi.org/10.3390/v13102015
- Jambunathan N., Clark C.M., Musarrat F., Chouljenko V.N., Rudd J., Kousoulas K.G. Two sides to every story: herpes simplex type-1 viral glycoproteins gB, gD, gH/gL, gK, and cellular receptors function as key players in membrane fusion. Viruses. 2021; 13(9): 1849. https://doi.org/10.3390/v13091849
- Rivas T., Goodrich J.A., Kugel J.F. The herpes simplex virus 1 protein ICP4 acts as both an activator and a repressor of host genome transcription during infection. Mol. Cell. Biol. 2021; 41(10): e0017121. https://doi.org/10.1128/MCB.00171-21
- Ding X., Neumann D.M., Zhu L. Host factors associated with either VP16 or VP16-induced complex differentially affect HSV-1 lytic infection. Rev. Med. Virol. 2022; 32(6): e2394. https://doi.org/10.1002/rmv.2394
- Sosnovtseva A.O., Demidova N.A., Klimova R.R., Kovalev M.A., Kushch A.A., Starodubova E.S., et al. Control of HSV-1 infection: directions for the development of CRISPR/Cas-Based therapeutics and diagnostics. Int. J. Mol. Sci. 2024; 25(22): 12346. https://doi.org/10.3390/ijms252212346
- Jiang S., Li H., Zhang L., Mu W., Zhang Y., Chen T., et al. Generic Diagramming Platform (GDP): a comprehensive database of high-quality biomedical graphics. Nucleic Acids Res. 2025; 53(D1): D1670–6. https://doi.org/10.1093/nar/gkae973
- Fox H.L., Dembowski J.A., DeLuca N.A. A herpesviral immediate early protein promotes transcription elongation of viral transcripts. mBio. 2017; 8(3): e00745-17. https://doi.org/10.1128/mBio.00745-17
- Renner D.W., Szpara M.L. Impacts of genome-wide analyses on our understanding of human herpesvirus diversity and evolution. J. Virol. 2018; 92(1): e00908-17. https://doi.org/10.1128/JVI.00908-17
- Grady L.M., Szczepaniak R., Murelli R.P., Masaoka T., Le Grice S.F.J., Wright D.L., et al. The exonuclease activity of herpes simplex virus 1 UL12 is required for production of viral DNA that can be packaged to produce infectious virus. J. Virol. 2017; 91(23): e01380-17. https://doi.org/10.1128/JVI.01380-17
- Waisner H., Lasnier S., Suma S.M., Kalamvoki M. Effects on exocytosis by two HSV-1 mutants unable to block autophagy. J. Virol. 2023; 97(10): e0075723. https://doi.org/10.1128/jvi.00757-23
- Suzich J.B., Cliffe A.R. Strength in diversity: Understanding the pathways to herpes simplex virus reactivation. Virology. 2018; 522: 81. https://doi.org/10.1016/j.virol.2018.07.011
- Nicoll M.P., Hann W., Shivkumar M., Harman L.E., Connor V., Coleman H.M., et al. The HSV-1 latency-associated transcript functions to repress latent phase lytic gene expression and suppress virus reactivation from latently infected neurons. PLoS Pathog. 2016; 12(4): e1005539. https://doi.org/10.1371/journal.ppat.1005539
- Sawtell N.M., Thompson R.L. De novo herpes simplex virus VP16 expression gates a dynamic programmatic transition and sets the latent/lytic balance during acute infection in trigeminal ganglia. PLoS Pathog. 2016; 12(9): e1005877. https://doi.org/10.1371/journal.ppat.1005877
- Cuddy S.R., Schinlever A.R., Dochnal S., Seegren P.V., Suzich J., Kundu P., et al. Neuronal hyperexcitability is a DLK-dependent trigger of herpes simplex virus reactivation that can be induced by IL-1. Elife. 2020; 9: e58037. https://doi.org/10.7554/eLife.58037
- Kostyusheva A., Brezgin S., Babin Y., Vasilyeva I., Glebe D., Kostyushev D., et al. CRISPR-Cas systems for diagnosing infectious diseases. Methods. 2022; 203: 431–46. https://doi.org/10.1016/j.ymeth.2021.04.007
- Freije C.A., Sabeti P.C. Detect and destroy: CRISPR-based technologies for the response against viruses. Cell Host Microbe. 2021; 29(5): 689–703. https://doi.org/10.1016/j.chom.2021.04.003
- Bairqdar A., Karitskaya P.E., Stepanov G.A. Expanding horizons of CRISPR/Cas technology: clinical advancements, therapeutic applications, and challenges in gene therapy. Int. J. Mol. Sci. 2024; 25(24): 13321. https://doi.org/10.3390/ijms252413321
- Boonbanjong P., Treerattrakoon K., Waiwinya W., Pitikultham P., Japrung D. Isothermal amplification technology for disease diagnosis. Biosensors (Basel). 2022; 12(9): 677. https://doi.org/10.3390/bios12090677
- Liu D.N., Wu H.P., Zhou G.H. Research progress of visual detection in rapid on-site detection of pathogen nucleic acid. Yi Chuan. 2023; 45(4): 306–23. https://doi.org/10.16288/j.yczz.22-323
- Mao X., Xu M., Luo S., Yang Y., Zhong J., Zhou J., et al. Advancements in the synergy of isothermal amplification and CRISPR-cas technologies for pathogen detection. Front. Bioeng. Biotechnol. 2023; 11: 1273988. https://doi.org/10.3389/fbioe.2023.1273988
- Volkov A., Dolgova A., Dedkov V. CRISPR/Cas-based diagnostic platforms. Infektsiya i immunitet. 2022; 12(1): 9–20. https://doi.org/10.15789/2220-7619-CCB-1843 https://elibrary.ru/fdhjgz (in Russian)
- Geojith G., Dhanasekaran S., Chandran S.P., Kenneth J. Efficacy of loop mediated isothermal amplification (LAMP) assay for the laboratory identification of Mycobacterium tuberculosis isolates in a resource limited setting. J. Microbiol. Methods. 2011; 84(1): 71–3. https://doi.org/10.1016/j.mimet.2010.10.015
- Uttam I., Sudarsan S., Ray R., Chinnappan R., Yaqinuddin A., Al-Kattan K., et al. A hypothetical approach to concentrate microorganisms from human urine samples using paper-based adsorbents for point-of-care molecular assays. Life (Basel). 2023; 14(1): 38. https://doi.org/10.3390/life14010038
- Lobato I.M., O’Sullivan C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Analyt. Chem. 2018; 98: 19–35. https://doi.org/10.1016/j.trac.2017.10.015
- Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016; 85: 227. https://doi.org/10.1146/annurev-biochem-060815-014607
- Chen Y., Zhao R., Hu X., Wang X. The current status and future prospects of CRISPR-based detection of monkeypox virus: A review. Anal. Chim. Acta. 2025; 1336: 343295. https://doi.org/10.1016/j.aca.2024.343295
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017; 356(6336): 438. https://doi.org/10.1126/science.aam9321
- Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018; 360(6387): 444–8. https://doi.org/10.1126/science.aas8836
- Srivastava S., Upadhyay D.J., Srivastava A. Next-generation molecular diagnostics development by CRISPR/Cas tool: rapid detection and surveillance of viral disease outbreaks. Front. Mol. Biosci. 2020; 7: 582499. https://doi.org/10.3389/fmolb.2020.582499
- Shi P., Wu X. Programmable RNA targeting with CRISPR-Cas13. RNA Biol. 2024; 21(1): 1–9. https://doi.org/10.1080/15476286.2024.2351657
- Calvert A.E., Biggerstaff B.J., Tanner N.A., Lauterbach M., Lanciotti R.S. Rapid colorimetric detection of Zika virus from serum and urine specimens by reverse transcription loop-mediated isothermal amplification (RT-LAMP). PLoS One. 2017; 12(9): e0185340. https://doi.org/10.1371/journal.pone.0185340
- Hang X.M., Liu P.F., Tian S., Wang H.Y., Zhao K.R., Wang L. Rapid and sensitive detection of Ebola RNA in an unamplified sample based on CRISPR-Cas13a and DNA roller machine. Biosens. Bioelectron. 2022; 211: 114393. https://doi.org/10.1016/j.bios.2022.114393
- Niu M., Dong Z., Yu L., Dong X., An J., Han Y., et al. Anti-RNA virus crRNA targets efficient screening platform based on bioinformatics and CRISPR detection. Mol. Ther. Nucleic Acids. 2025; 36(3): 102619. https://doi.org/10.1016/j.omtn.2025.102619
- Saju A.F., Mukundan A., Divyashree M., Chandrashekhar R., Mahadev Rao A. RNA diagnostics and therapeutics: a comprehensive review. RNA Biol. 2025; 22(1): 1–11. https://doi.org/10.1080/15476286.2024.2449277
- Cherkaoui D., Huang D., Miller B.S., Turbe V., McKendry R.A. Harnessing recombinase polymerase amplification for rapid multi-gene detection of SARS-CoV-2 in resource-limited settings. Biosens. Bioelectron. 2021; 189: 113328. https://doi.org/10.1016/j.bios.2021.113328
- Ramadan N.K., Gaber N., Ali N.M., Amer O.S.O., Soliman H. SHERLOCK, a novel CRISPR-Cas13a-based assay for detection of infectious bursal disease virus. J. Virol. Methods. 2025; 337: 115185. https://doi.org/10.1016/j.jviromet.2025.115185
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018; 360(6387): 436. https://doi.org/10.1126/science.aar6245
- Li L., Li S., Wu N., Wu J., Wang G., Zhao G., et al. HOLMESv2: A CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019; 8(10): 2228–37. https://doi.org/10.1021/acssynbio.9b00209
- Zhuang S., Hu T., Zhou H., He S., Li J., Zhang Y., et al. CRISPR-HOLMES-based NAD(+) detection. Front. Bioeng. Biotechnol. 2024; 12: 1355640. https://doi.org/10.3389/fbioe.2024.1355640
- Yang D., Shi Y., Tang Y., Yin H., Guo Y., Wen S., et al. Effect of HPV infection on the occurrence and development of laryngeal cancer: a review. J. Cancer. 2019; 10(19): 4455–62. https://doi.org/10.7150/jca.34016
- Yin L., Zhao Z., Wang C., Zhou C., Wu X., Gao B., et al. Development and evaluation of a CRISPR/Cas12a-based diagnostic test for rapid detection and genotyping of HR-HPV in clinical specimens. Microbiol. Spectr. 2025; 13(1): e0225324. https://doi.org/10.1128/spectrum.02253-24
- Wu X., Chan C., Springs S.L., Lee Y.H., Lu T.K., Yu H. A warm-start digital CRISPR/Cas-based method for the quantitative detection of nucleic acids. Anal. Chim. Acta. 2022; 1196: 339494. https://doi.org/10.1016/j.aca.2022.339494
- Yin X., Luo H., Zhou H., Zhang Z., Lan Y., Feng Z., et al. A rapid isothermal CRISPR-Cas13a diagnostic test for genital herpes simplex virus infection. iScience. 2024; 27(1): 108581. https://doi.org/10.1016/j.isci.2023.108581
- Huang M., Chen Y., Zheng L., Yao Y.F. Highly sensitive and naked-eye detection of herpes simplex virus type 1 using LAMP-CRISPR/Cas12 diagnostic technology and gold nanoparticles. Heliyon. 2023; 9(11): e22146. https://doi.org/10.1016/j.heliyon.2023.e22146
- FDA Approves first gene therapies to treat patients with sickle cell disease. Available at: https://fda.gov/news-events/press-announcements/fda-approves-first-gene-therapies-treat-patients-sickle-cell-disease
- Bellizzi A., Cakir S., Donadoni M., Sariyer R., Liao S., Liu H., et al. Suppression of HSV-1 infection and viral reactivation by CRISPR-Cas9 gene editing in 2D and 3D culture models. Mol. Ther. Nucleic Acids. 2024; 35(3): 102282. https://doi.org/10.1016/j.omtn.2024.102282
- Katson M., Gorenshtein A., Pepys J., Mina Y., Shelly S. Mortality and prognosis in herpes simplex Virus-1 encephalitis long-term follow up study. J. Neurol. Sci. 2025; 468: 123330. https://doi.org/10.1016/j.jns.2024.123330
- Ying M., Wang H., Liu T., Han Z., Lin K., Shi Q., et al. CLEAR strategy inhibited HSV proliferation using viral vectors delivered CRISPR-Cas9. Pathogens. 2023; 12(6): 814. https://doi.org/10.3390/pathogens12060814
- McCormick I., James C., Welton N.J., Mayaud P., Turner K.M.E., Gottlieb S.L., et al. Incidence of herpes simplex virus keratitis and other ocular disease: global review and estimates. Ophthalmic Epidemiol. 2022; 29(4): 353–62. https://doi.org/10.1080/09286586.2021.1962919
- Yin D., Ling S., Wang D., Dai Y., Jiang H., Zhou X., et al. Targeting herpes simplex virus with CRISPR-Cas9 cures herpetic stromal keratitis in mice. Nat. Biotechnol. 2021; 39(5): 567–77. https://doi.org/10.1038/s41587-020-00781-8
- Amrani N., Luk K., Singh P., Shipley M., Isik M., Donadoni M., et al. CRISPR-Cas9-mediated genome editing delivered by a single AAV9 vector inhibits HSV-1 reactivation in a latent rabbit keratitis model. Mol. Ther. Methods Clin. Dev. 2024; 32(3): 101303. https://doi.org/10.1016/j.omtm.2024.101303
- Dutton A.J., Turnbaugh E.M., Patel C.D., Garland C.R., Taylor S.A., Alers-Velazquez R., et al. Asymptomatic neonatal herpes simplex virus infection in mice leads to persistent CNS infection and long-term cognitive impairment. PLoS Pathog. 2025; 21(2): e1012935. https://doi.org/10.1371/journal.ppat.1012935
- Wei A., Yin D., Zhai Z., Ling S., Le H., Tian L., et al. In vivo CRISPR gene editing in patients with herpetic stromal keratitis. Mol. Ther. 2023; 31(11): 3163–75. https://doi.org/10.1016/j.ymthe.2023.08.021
- ClinicalTrials.gov. A Study of the Safety, Tolerability and Prelinminary Efficacy of BD111 in Herpes Simplex Virus Type I Stromal Keratitis; 2025. Available at: https://clinicaltrials.gov/study/NCT06474416
Supplementary files