The molecular evolution of Dabie bandavirus (Phenuiviridae: Bandavirus: Dabie bandavirus), the agent of severe fever with thrombocytopenia syndrome
- Authors: Sizikova T.E.1, Lebedev V.N.1, Borisevich S.V.1
-
Affiliations:
- FSBI «Central Scientific Research Institute No. 48» of the Ministry of Defense of Russian Federation
- Issue: Vol 66, No 6 (2021)
- Pages: 409-416
- Section: REVIEWS
- URL: https://ogarev-online.ru/0507-4088/article/view/118195
- DOI: https://doi.org/10.36233/0507-4088-68
- ID: 118195
Cite item
Full Text
Abstract
Since the Dabie bandavirus (DBV; former SFTS virus, SFTSV) was identified, the epidemics of severe fever with thrombocytopenic syndrome (SFTS) caused by this virus have occurred in several countries in East Asia. The rapid increase in incidence indicates that this infectious agent has a pandemic potential and poses an imminent global public health threat.
The analysis of molecular evolution of SFTS agent that includes its variants isolated in China, Japan and South Korea was performed in this review. The evolution rate of DBV and the estimated dates of existence of the common ancestor were ascertained, and the possibility of reassortation was demonstrated.
The evolutionary rates of DBV genome segments were estimated to be 2.28 × 10-4 nucleotides/site/year for S-segment, 2.42 × 10-4 for M-segment, and 1.19 × 10-4 for L-segment. The positions of positive selection were detected in the viral genome.
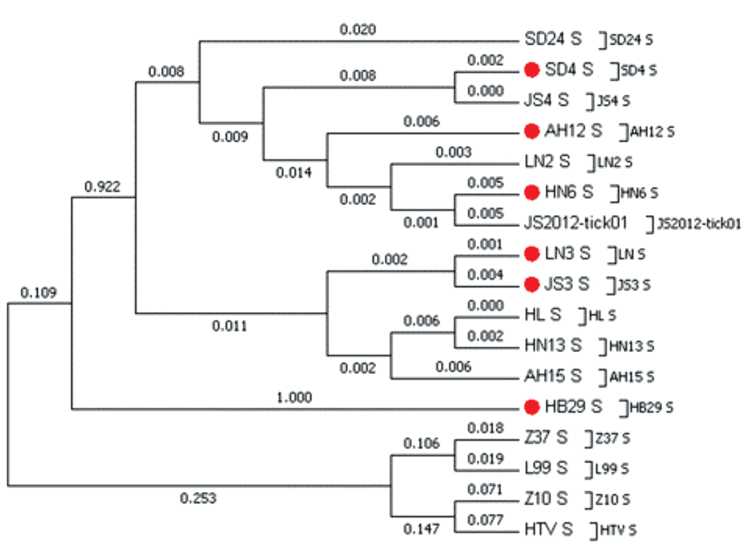
Phylogenetic analyses showed that virus may be divided into two clades, containing six different genotypes. The structures of phylogenetic trees for S-, M- and L-segments showed that all genotypes originate from the common ancestor.
Data of sequence analysis suggest that DBV use several mechanisms to maintain the high level of its genetic diversity. Understanding the phylogenetic factors that determine the virus transmission is important for assessing the epidemiological characteristics of the disease and predicting its possible outbreaks.
Full Text
##article.viewOnOriginalSite##About the authors
T. E. Sizikova
FSBI «Central Scientific Research Institute No. 48» of the Ministry of Defense of Russian Federation
ORCID iD: 0000-0002-1817-0126
141306, Sergiev Posad, Russia
Russian FederationV. N. Lebedev
FSBI «Central Scientific Research Institute No. 48» of the Ministry of Defense of Russian Federation
ORCID iD: 0000-0002-6552-4599
141306, Sergiev Posad, Russia
Russian FederationS. V. Borisevich
FSBI «Central Scientific Research Institute No. 48» of the Ministry of Defense of Russian Federation
Author for correspondence.
Email: 48cnii@mil.ru
ORCID iD: 0000-0002-6742-3919
Sergey V. Borisevich, D.Sci. (Biol.), Professor, Corresponding Member of the RAS, Director
141306, Sergiev Posad, Russia
Russian FederationReferences
- Kim K.H., Yi J., Kim G., Choi S.J., Jun K.I., Kim M.H., et al. Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg. Infect. Dis. 2013; 19(11): 1892–4. https://doi.org/10.3201/eid1911.130792
- Yu X.J., Liang M.F., Zhang S.Y., Liu Y., Li J.D., Sun Y.L., et al. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 2011; 364(16): 1523–32. https://doi.org/10.1056/NEJMoa1010095
- Zhao L., Zhai S., Wen H., Cui F., Chi Y., Wang L., et al. Severe fever with thrombocytopenia syndrome virus, Shandong Province, China. Emerg. Infect. Dis. 2012; 18(6): 963–5. https://doi.org/10.3201/eid1806.111345
- Niu G., Li J., Liang M., Jiang X., Jiang M., Yin H., et al. Severe fever with thrombocytopenia syndrome virus among domesticated animals, China. Emerg. Infect. Dis. 2013; 19(5): 756–63. https://doi.org/10.3201/eid1905.120245
- Takahashi T., Maeda K., Suzuki T., Ishido A., Shigeoka T., Tominaga T., et al. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J. Infect. Dis. 2014; 209(6): 816–27. https://doi.org/10.1093/infdis/jit6036. ICTV Taxonomy history: Dabie bandavirus. https://talk.ictvonline.org/taxonomy/p/taxonomy-history?taxnode_id=201900166 (accessed October 14, 2021).
- Lam T., Liu W., Bowden T.A., Gui N., Zhuang L., Liu K., et al. Evolutionary and molecular analysis of the emergent severe fever with thrombocytopenia syndrome virus. Epidemics. 2013; 5(1): 1–10. https://doi.org/10.1016/j.epidem.2012.09.002
- Xu B., Liu L., Huang X., Ma H., Zhang Y., Du Y., et al. Metagenomic Analysis of Fever, Thrombocytopenia and Leukopenia Syndrome (FTLS) in Henan Province, China: Discovery of a New Bunyavirus. PLoS Pathog. 2011; 7(11): e1002369. https://doi.org/10.1371/journal.ppat.1002369
- Yoshikawa T., Shimojima M., Fukushi S., Tani H., Fukuma A., Taniguchi S. Phylogenetic and geographic relationships of severe fever with thrombocytopenia syndrome virus in China, South Korea, and Japan. J. Infect. Dis. 2015; 212(6): 889–98. https://doi.org/10.1093/infdis/jiv144
- Zhang Y.Z., Zhou D.J., Qin X.C., Tian J.H., Xiong Y., Wang J.B., et al. The Ecology, Genetic Diversity, and Phylogeny of Huaiyangshan Virus in China. J. Virol. 2012; 86(5): 2864–8. https://doi.org/10.1128/JVI.06192-11
- Zhang Y.Z., Zhou D.J., Xiong Y. Hemorrhagic fever caused by a novel tick–borne Bunyavirus in Huaiynshan, China. Zhonghua Liu Xing Bing Xue Za Zhi (Chinese Journal of Epidemiology). 2011; 32(3):209–20. https://doi.org/10.3760/cma.j.issn.0254-6450.2011.03.001
- Yamanaka A., Kirino Y., Fujimoto S., Ueda N., Himeji D., Miura M., et al. Direct transmission of severe fever with thrombocytopenia syndrome virus from domestic cat to veterinary personnel. Emerg. Infect. Dis. 2020; 26(12): 2994–8. https://doi.org/10.3201/eid2612.191513
- Liu L., Chen W., Yang Y., Jiang Y. Molecular evolution of fever, thrombocytopenia and leukocytopenia virus (FTLSV) based on whole–genome sequences. Infect. Genet. Evol. 2016; 39: 55–63. https://doi.org/10.1016/j.meegid.2015.12.022
- Qu B., Qi X., Wu X., Liang M., Li C., Cardona C.J. Supression of the interferon and NF-κB responses by severe fever with thrombocytopenia syndrome virus. J. Virol. 2012; 86(16): 8388–401. https://doi.org/10.1128/JVI.00612-12
- Matsuno K., Weisend C., Travassos da Rosa A.P., Anzick S.L., Dahlstrom E., Porcella S.F., et al. Characterization of the Bhanja serogroup viruses (Bunyaviridae): a novel species of the genus Phlebovirus and its relationship with other emerging tick-borne phleboviruses. J. Virol. 2013; 87(7): 3719–28. https://doi.org/10.1128/JVI.02845-12
- Li A., Liu L., Wu W., Liu Y., Huang X., Li C., et al. Molecular evolution and genetic diversity analysis of SFTS virus based on next-generation sequencing. Biosaf. Health. 2021; 3(2): 105–15. https://doi.org/10.1016/j.bsheal.2021.02.002
- Huang X., Liu L., Du Y., Wu W., Wang H., Su J. The evolutionary history and spatiotemporal dynamics of the fever, thrombocytopenia and leukocytopenia syndrome virus (FTLSV) in China. PLoS Negl. Trop. Dis. 2014; 8(10): 1–13. https://doi.org/10.1371/journal.pntd.0003237
- McMullan L.K., Folk S.M., Kelly A.J., MacNeil A., Goldsmith C.S., Metcalfe M.G., et al. A new phlebovirus associated with severe febrile illness in Missouri. N. Engl. J. Med. 2012; 367(9):834–41. https://doi.org/10.1056/NEJMoa1203378
- Mourya D.T., Yadav P.D., Basu A., Shete A., Patil D.Y., Zawar D., et al. Malsoor Virus, a Novel Bat Phlebovirus, is Closely Related to Severe Fever with Thrombocytopenia Syndrome Virus and Heartland Virus. J. Virol. 2014; 88(6): 3605–9. https://doi.org/10.1128/JVI.02617-13
- Wang J., Selleck P., Yu M., Ha W., Rootes C., Gales R., et al. Novel phlebovirus with zoonotic potential isolated from ticks, Australia. Emerg. Infect. Dis. 2014; 20(6): 1040–3. https://doi.org/10.3201/eid2006.140003
- Chen X., Ye H., Li S., Jiao B., Wu J., Zeng P., et al. Severe fever with thrombocytopenia syndrome virus inhibits exogenous Type I IFN signaling pathway through its NSs in vitro. PloS One. 2017; 12(2): e0172744. https://doi.org/10.1371/journal.pone.0172744
- Liu L., Chen W., Yang Y., Jiang Y. Molecular evolution of fever, thrombocytopenia and leukocytopenia virus (FTLSV) based on whole-genome sequences. Infect. Genet. Evol. 2016; 39: 55–63. https://doi.org/10.1016/j.meegid.2015.12.022
- Kosakovsky P.S.L., Poon A.F., Leigh B.A.J., Frost S.D. A maximum likelihood method for detecting directional evolution in protein sequences and its application to influenza A virus. Mol. Biol. Evol. 2008; 25(9): 1809–24. https://doi.org/10.1093/molbev/msn123
- Lu S., Wang L., Bai D., Li U. Establishment of national reference for bunyavirus nucleic acid detection kits for diagnosis of SFTS virus. Virol. J. 2017; 14(1): 32. https://doi.org/10.1186/s12985-017-0682-z
- Liu J.W., Zhao L., Luo L.M., Liu M.M., Sun Y., Su X., et al. Molecular evolution and spatial transmission of severe fever with thrombocytopenia syndrome virus based on complete genome sequences. PLoS One. 2016; 11(3): e0151677. https://doi.org/10.1371/journal.pone.0151677
- Feng C., Zhang L., Sun Y., Shao B., Mao H., Jiang J., et al. Genome sequencing and the molecular evolutionary analysis of a SFTSV isolated from Zhejiang province. Zhonghua Yu Fang Yi Xue Za Zhi (Chinese journal of preventive medicine). 2014; 48(7): 612–6. https://doi.org/10.3760/cma.j.issn.0253-9624.2014.07.016 (in Chinese)
- Ding N.Z., Luo Z.F., Niu D.D., Ji W., Kang X.H., Cai S.S., et al. Identification of two severe fever with thrombocytopenia syndrome virus strains originating from reassortment. Virus Res. 2013; 178(2):543–6. https://doi.org/10.1016/j.virusres.2013.09.017
- Fu Y., Li S., Zhang Z., Man S., Li X., Zhang W. Phylogeographic analysis of severe fever with thrombocytopenia syndrome virus from Zhoushan Islands, China: implication for transmission across the ocean. Sci. Rep. 2016; 6: 19563. https://doi.org/10.1038/srep19563
- Bowen M.D., Trappier S.G., Sanchez A.J., Meyer R.F., Goldsmith C.S., Zaki S.R., et al. A reassortant bunyavirus isolated from acute hemorrhagic fever cases in Kenya and Somalia. Virology. 2001; 291(2): 185–90. https://doi.org/10.1006/viro.2001.1201
- Briese T., Bird B., Kapoor V., Nichol S.T., Lipkin W.I. Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa. J. Virol. 2006; 80(11):5627–30. https://doi.org/10.1128/jvi.02448-05
- Chandler L.J., Hogge G., Endres M., Jacoby D.R., Nathanson N., Beaty B.J. Reassortment of La Crosse and Tahyna bunyaviruses in Aedes triseriatus mosquitoes. Virus Res. 1991; 20(2): 181–91. https://doi.org/10.1016/0168-1702(91)90108-8
- Gerrard S.R., Li L., Barrett A.D., Nichol S.T. Ngari virus is a Bunyamwera virus reassortant that can be associated with large outbreaks of hemorrhagic fever in Africa. J. Virol. 2004; 78(16):8922–6. https://doi.org/10.1128/jvi.78.16.8922-8926.2004
- Saeed M.F., Wang H., Suderman M., Beasley D.W., Travassos da Rosa A., Li L., et al. Jatobal virus is a reassortant containing the small RNA of Oropouche virus. Virus Res. 2001; 77(1): 25–30. https://doi.org/10.1016/s0168-1702(01)00262-3
- Deadly new flu virus in US and Mexico may go pandemic. https://www.newscientist.com/article/dn17025-deadly-new-flu-virus-in-usand-mexico-may-go-pandemic/?ignored=irrelevant (accessed October 14, 2021).
- Ding F., Zhang W., Wang L., Hu W., Soares Magalhaes R.J., Sun H., et al. Epidemiologic features of severe fever with thrombocytopenia syndrome in China, 2011–2012. Clin. Infect. Dis. 2013; 56(11):1682–3. https://doi.org/10.1093/cid/cit100
- Alberts B., Bray D., Roberts K., Lewis J., Raff M. Essential Cell Biology: An Introduction to the Molecular Biology of the Cell. London: Taylor & Francis; 1997.
- Casel M.A., Park S.J., Choi Y.K. Severe fever with thrombocytopenia syndrome virus: emerging novel phlebovirus and their control strategy. Exp. Mol. Med. 2021; 53: 713–22. https://doi.org/10.1038/s12276-021-00610-1
- Sato Y., Mekata H., Sudaryatma P.E., Kirino Y., Yamamoto S., Ando S., et al. Isolation of severe fever with thrombocytopenia syndrome virus from various tick species in area with human severe fever with thrombocytopenia syndrome cases. Vector Borne Zoonotic Dis. 2021; 21(5): 378–84. https://doi.org/10.1089/vbz.2020.2720
Supplementary files