Генетическое разнообразие белка Vif у вариантов вируса иммунодефицита человека 1-го типа (Retroviridae: Orthoretrovirinae: Lentivirus: Human immunodeficiency virus-1), циркулировавших в Московской области в 2019‒2020 гг.
- Авторы: Антонова А.А.1, Протасова Л.А.1, Ким К.В.1, Мунчак Я.М.1, Меженская Е.Н.1, Орлова-Морозова Е.А.2, Пронин А.Ю.2, Прилипов А.Г.1, Кузнецова А.И.1
-
Учреждения:
- Институт вирусологии им. Д.И. Ивановского ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи» Минздрава России
- ГБУЗ Московской области «Центр профилактики и борьбы со СПИД»
- Выпуск: Том 70, № 2 (2025)
- Страницы: 117-132
- Раздел: ОРИГИНАЛЬНЫЕ ИССЛЕДОВАНИЯ
- URL: https://ogarev-online.ru/0507-4088/article/view/310651
- DOI: https://doi.org/10.36233/0507-4088-281
- EDN: https://elibrary.ru/qoqqce
- ID: 310651
Цитировать
Аннотация
Введение. Белок Vif вируса иммунодефицита человека 1-го типа (ВИЧ-1) является фактором вирусной инфекционности и противодействует клеточным дезаминазам семейства APOBEC3, препятствующим репликации вируса. На основе белка Vif ведутся разработки для создания терапевтических средств. Природные замены в белке Vif могут влиять на его функциональность и ассоциироваться с ускоренным переходом ВИЧ-инфекции в стадию СПИДа. Изучение особенностей белка Vif у вариантов ВИЧ-1, циркулирующих в России, ранее не проводилось.
Цель работы: изучить генетическое разнообразие белка Vif вариантов ВИЧ-1, циркулировавших в Московской области в 2019‒2020 гг.
Материалы и методы. Проанализировано 234 образца цельной крови ВИЧ-инфицированных пациентов без опыта приема антиретровирусной терапии. Дизайн исследования включал следующие стадии: экстракция провирусной ДНК, амплификация гена vif, секвенирование продуктов амплификации, определение генетических вариантов полученных нуклеотидных последовательностей; затем проводили исследование консенсусных последовательностей белка Vif наиболее распространенных генетических вариантов ВИЧ-1, анализ консервативности и генетического разнообразия белка Vif-А6 (белок Vif вариантов ВИЧ-1 суб-субтипа А6) у пациентов с разными стадиями заболевания, оценку генетического разнобразия белка Vif-А6 в Московской области.
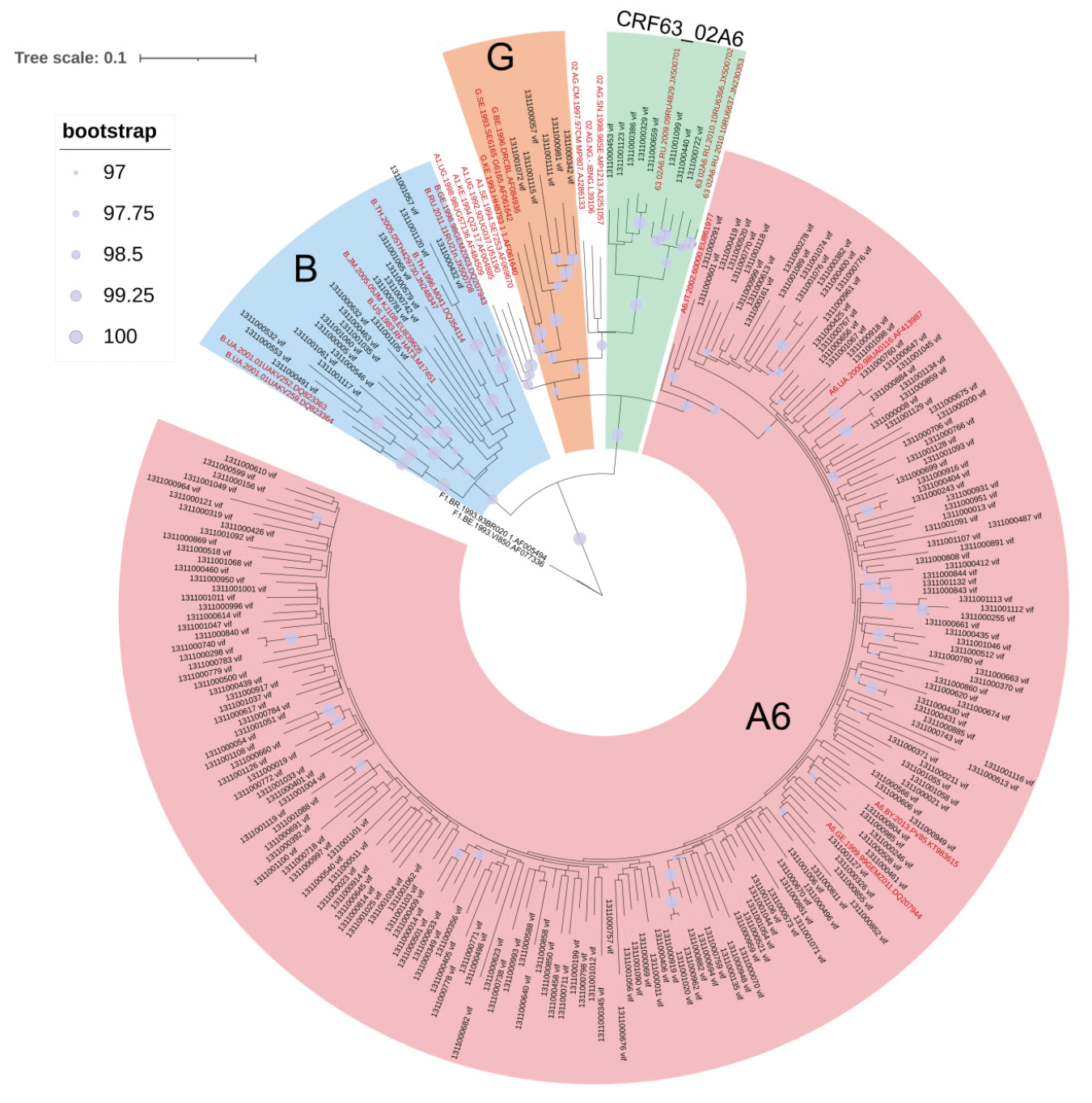
Результаты. Проведена оценка генетического разнообразия ВИЧ-1 в области генома, кодирующего белок Vif, – выявлено большое генетическое разнообразие, включающее в себя ВИЧ-1 чистых субтипов (A6, B и G), а также его рекомбинантных форм (CRF63_02A6). Впервые получены консенсусные последовательности белка Vif генетических вариантов ВИЧ-1 – B и CRF63_02A6, циркулирующих на территории Российской Федерации. Для наиболее распространенных в России вариантов ВИЧ-1 определены характерные замены в консенсусных последовательностях.
Заключение. Ограничением исследования является небольшая выборка последовательностей, принадлежащих генетическим вариантам B и CRF63_02A6 ВИЧ-1. Полученные результаты могут быть интересны и учтены при разработке терапевтических средств на основе белка Vif, а также при изучении вопросов патогенности ВИЧ-1 суб-субтипа А6.
Ключевые слова
Полный текст
Открыть статью на сайте журналаОб авторах
Анастасия Александровна Антонова
Институт вирусологии им. Д.И. Ивановского ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи» Минздрава России
Email: anastaseika95@mail.ru
ORCID iD: 0000-0002-9180-9846
канд. биол. наук, научный сотрудник лаборатории вирусов лейкозов
Россия, 123098, г. МоскваЛариса Анатольевна Протасова
Институт вирусологии им. Д.И. Ивановского ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи» Минздрава России
Email: larisa.protasova.03@mail.ru
ORCID iD: 0009-0001-0430-1578
лаборант-исследователь лаборатории вирусов лейкозов
Россия, 123098, г. МоскваКристина Вячеславовна Ким
Институт вирусологии им. Д.И. Ивановского ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи» Минздрава России
Email: kimsya99@gmail.com
ORCID iD: 0000-0002-4150-2280
младший научных сотрудник лаборатории вирусов лейкозов
Россия, 123098, г. МоскваЯна Михайловна Мунчак
Институт вирусологии им. Д.И. Ивановского ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи» Минздрава России
Email: yana_munchak@mail.ru
ORCID iD: 0000-0002-4792-8928
младший научный сотрудник лаборатории вирусов лейкозов
Россия, 123098, г. МоскваЕкатерина Никитична Меженская
Институт вирусологии им. Д.И. Ивановского ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи» Минздрава России
Email: belokopytova.01@mail.ru
ORCID iD: 0000-0002-3110-0843
канд. биол. наук, научный сотрудник лаборатории вирусов лейкозов
Россия, 123098, г. МоскваЕлена Александровна Орлова-Морозова
ГБУЗ Московской области «Центр профилактики и борьбы со СПИД»
Email: orlovamorozova@gmail.com
ORCID iD: 0000-0003-2495-6501
канд. мед. наук, заведующая амбулаторно-поликлиническим отделением
Россия, 140053, Московская область, г. КотельникиАлександр Юрьевич Пронин
ГБУЗ Московской области «Центр профилактики и борьбы со СПИД»
Email: alexanderp909@gmail.com
ORCID iD: 0000-0001-9268-4929
канд. мед. наук, главный врач
Россия, 140053, Московская область, г. КотельникиАлексей Геннадьевич Прилипов
Институт вирусологии им. Д.И. Ивановского ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи» Минздрава России
Email: a_prilipov@mail.ru
ORCID iD: 0000-0001-8755-1419
д-р биол. наук, ведущий научный сотрудник, заведующий лабораторией молекулярной генетики
Россия, 123098, г. МоскваАнна Игоревна Кузнецова
Институт вирусологии им. Д.И. Ивановского ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи» Минздрава России
Автор, ответственный за переписку.
Email: a-myznikova@list.ru
ORCID iD: 0000-0001-5299-3081
заведующая лаборатории вирусов лейкозов, канд. биол. наук, ведущий научный сотрудник
Россия, 123098, г. МоскваСписок литературы
- Rose K.M., Marin M., Kozak S.L., Kabat D. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 2004; 10(6): 291–7. https://doi.org/10.1016/j.molmed.2004.04.008
- Stupfler B., Verriez C., Gallois-Montbrun S., Marquet R., Paillart J.C. Degradation Independent inhibition of APOBEC3G by the HIV-1 Vif protein. Viruses. 2021; 13(4): 617. https://doi.org/10.3390/v13040617
- Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002; 418(6898): 646–50. https://doi.org/10.1038/nature00939
- Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003; 424(6944): 99–103. https://doi.org/10.1038/nature01709
- Hultquist J.F., Lengyel J.A., Refsland E.W., LaRue R.S., Lackey L., Brown W.L., et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-Deficient HIV-1. J. Virol. 2011; 85(21): 11220–34. https://doi.org/10.1128/JVI.05238-11
- Wang X., Abudu A., Son S., Dang Y., Venta P.J., Zheng Y.H. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J. Virol. 2011; 85(7): 3142–52. https://doi.org/10.1128/JVI.02049-10
- Guo F., Cen S., Niu M., Yang Y., Gorelick R.J., Kleiman L. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J. Virol. 2007;81(20):11322–31. https://doi.org/10.1128/JVI.00162-07
- Xu W.K., Byun H., Dudley J.P. The role of APOBECs in viral replication. Microorganisms. 2020; 8(12): 1899. https://doi.org/10.3390/microorganisms8121899
- Azimi F.C., Lee J.E. Structural perspectives on HIV-1 Vif and APOBEC3 restriction factor interactions. Protein Sci. 2020; 29(2): 391–406. https://doi.org/10.1002/pro.3729
- Friedler A., Zakai N., Karni O., Friedler D., Gilon C., Loyter A. Identification of a nuclear transport inhibitory signal (NTIS) in the basic domain of HIV-1 Vif protein. J. Mol. Biol. 1999; 289(3): 431–7. https://doi.org/10.1006/jmbi.1999.2785
- Takaori-Kondo A., Shindo K. HIV-1 Vif: a guardian of the virus that opens up a new era in the research field of restriction factors. Front. Microbiol. 2013; 4: 34. https://doi.org/10.3389/fmicb.2013.00034
- Simon V., Zennou V., Murray D., Huang Y., Ho D.D., Bieniasz P.D. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005; 1(1): e6. https://doi.org/10.1371/journal.ppat.0010006
- Iwabu Y., Kinomoto M., Tatsumi M., Fujita H., Shimura M., Tanaka Y., et al. Differential anti-APOBEC3G activity of HIV-1 Vif proteins derived from different subtypes. J. Biol. Chem. 2010; 285(46): 35350–8. https://doi.org/10.1074/jbc.M110.173286
- Ronsard L., Raja R., Panwar V., Saini S., Mohankumar K., Sridharan S., et al. Genetic and functional characterization of HIV-1 Vif on APOBEC3G degradation: First report of emergence of B/C recombinants from North India. Sci. Rep. 2015; 5: 15438. https://doi.org/10.1038/srep15438
- Громов К.Б., Лага В.Ю., Мурзакова А.В., Киреев Д.Е. Анализ полиморфизма неструктурных генов ВИЧ-1 Vif и Rev. В кн.: Молекулярная диагностика – 2017: сборник трудов IХ Всероссийской научно-практической конференции с международным участием. М.; 2017: 455–6.
- De Maio F.A., Rocco C.A., Aulicino P.C., Bologna R., Mangano A., Sen L. Effect of HIV-1 Vif variability on progression to pediatric AIDS and its association with APOBEC3G and CUL5 polymorphisms. Infect. Genet. Evol. 2011; 11(6): 1256–62. https://doi.org/10.1016/j.meegid.2011.04.020
- Bizinoto M.C., Yabe S., Leal É., Kishino H., Martins L. de O., de Lima M.L., et al. Codon pairs of the HIV-1 vif gene correlate with CD4+ T cell count. BMC Infect. Dis. 2013; 13: 173. https://doi.org/10.1186/1471-2334-13-173
- Villanova F., Barreiros M., Janini L.M., Diaz R.S., Leal É. Genetic diversity of HIV-1 gene Vif among treatment-naive Brazilians. AIDS Res. Hum. Retroviruses. 2017; 33(9): 952–9. https://doi.org/10.1089/AID.2016.0230
- Bennett R.P., Salter J.D., Smith H.C. A new class of antiretroviral enabling innate immunity by protecting APOBEC3 from HIV Vif-dependent degradation. Trends Mol. Med. 2018; 24(5): 507–20. https://doi.org/10.1016/j.molmed.2018.03.004
- Sharkey M., Sharova N., Mohammed I., Huff S.E., Kummetha I.R., Singh G., et al. HIV-1 escape from small-molecule antagonism of Vif. mBio. 2019; 10(1): e00144-19. https://doi.org/10.1128/mBio.00144-19
- Duan S., Wang S., Song Y., Gao N., Meng L., Gai Y., et al. A novel HIV-1 inhibitor that blocks viral replication and rescues APOBEC3s by interrupting Vif/CBFβ interaction. J. Biol. Chem. 2020; 295(43): 14592–605. https://doi.org/10.1074/jbc.RA120.013404
- Akbari E., Seyedinkhorasani M., Bolhassani A. Conserved multiepitope vaccine constructs: A potent HIV-1 therapeutic vaccine in clinical trials. Braz. J. Infect. Dis. 2023; 27(3): 102774. https://doi.org/10.1016/j.bjid.2023.102774
- Guerra-Palomares S.E., Hernandez-Sanchez P.G., Esparza-Perez M.A., Arguello J.R., Noyola D.E., Garcia-Sepulveda C.A. Molecular characterization of Mexican HIV-1 Vif sequences. AIDS Res. Hum. Retroviruses. 2016; 32(3): 290–5. https://doi.org/10.1089/AID.2015.0290
- Bbosa N., Kaleebu P., Ssemwanga D. HIV subtype diversity worldwide. Curr. Opin. HIV AIDS. 2019; 14(3): 153–60. https://doi.org/10.1097/COH.0000000000000534
- Williams M.E. HIV-1 Vif protein sequence variations in South African people living with HIV and their influence on Vif-APOBEC3G interaction. Eur. J. Clin. Microbiol. Infect. Dis. 2024; 43(2): 325–38. https://doi.org/10.1007/s10096-023-04728-0
- Антонова А.А., Кузнецова А.И., Ожмегова Е.Н., Лебедев А.В., Казеннова Е.В., Ким К.В. и др. Генетическое разнообразие ВИЧ-1 на современном этапе эпидемии в Российской Федерации: увеличение распространенности рекомбинантных форм. ВИЧ-инфекция и иммуносупрессии. 2023; 15(3): 61–72. https://doi.org/10.22328/2077-9828-2023-15-3-61-72 https://elibrary.ru/tpwttn
- Кузнецова А.И., Громов К.Б., Киреев Д.Е., Шлыкова А.В., Лопатухин А.Э., Казеннова Е.В. и др. Анализ особенностей белка Tat вируса иммунодефицита человека 1 типа суб-субтипа А6 (Retroviridae: Orthoretrovirinae: Lentivirus: Human immunodeficiency virus-1). Вопросы вирусологии. 2021; 66(6): 452–63. https://doi.org/10.36233/0507-4088-83 https://elibrary.ru/cmzgyc
- Kuznetsova A., Kim K., Tumanov A., Munchak I., Antonova A., Lebedev A., et al. Features of Tat protein in HIV-1 sub-subtype A6 variants circulating in the Moscow Region, Russia. Viruses. 2023; 15(11): 2212. https://doi.org/10.3390/v15112212 https://elibrary.ru/ucqyal
- Антонова А.А., Лебедев А.В., Казеннова Е.В., Ким К.В., Ожмегова Е.Н., Туманов А.С. и др. Вариабельность белка VPU ВИЧ-1 суб-субтипа А6 у пациентов c различными стадиями ВИЧ-инфекции. ВИЧ-инфекция и иммуносупрессии. 2024; 16(2): 40–50. https://doi.org/10.22328/2077-9828-2024-16-2-40-50 https://elibrary.ru/lpjxqk
- Lebedev A., Kim K., Ozhmegova E., Antonova A., Kazennova E., Tumanov A., et al. Rev protein diversity in HIV-1 group M clades. Viruses. 2024; 16(5): 759. https://doi.org/10.3390/v16050759
- Антонова А.А., Лебедев А.В., Ожмегова Е.Н., Шлыкова А.В., Лаповок И.А., Кузнецова А.И. Вариабельность неструктурных белков у вариантов ВИЧ-1 суб-субтипа А6 (Retroviridae: Orthoretrovirinae: Lentivirus: Human immunodeficiency virus-1, sub-subtype A6), циркулирующих в разных регионах Российской Федерации. Вопросы вирусологии. 2024; 69(5): 470–80. https://doi.org/10.36233/0507-4088-262 https://elibrary.ru/wbbkuq
- Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic. Acids. Res. 1988; 16(3): 1215. https://doi.org/10.1093/nar/16.3.1
- Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014; 30(22): 3276–8. https://doi.org/10.1093/bioinformatics/btu531
- Struck D., Lawyer G., Ternes A.M., Schmit J.C., Bercoff D.P. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res. 2014; 42(18): e144. https://doi.org/10.1093/nar/gku739
- Schultz A.K., Bulla I., Abdou-Chekaraou M., Gordien E., Morgenstern B., Zoaulim F., et al. jpHMM: recombination analysis in viruses with circular genomes such as the hepatitis B virus. Nucleic Acids Res. 2012; 40: W193-8. https://doi.org/10.1093/nar/gks414.
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015; 32(1): 268–74. https://doi.org/10.1093/molbev/msu300
- Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012; 9(8): 772. https://doi.org/10.1038/nmeth.2109.
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021; 49(W1): W293–6. https://doi.org/10.1093/nar/gkab301
- Березов Т.Т., Коровкин Б.Ф. Биологическая химия. М.: Медицина; 1998.
- Lobanov M.Y., Pereyaslavets L.B., Likhachev I.V., Matkarimov B.T., Galzitskaya O.V. Is there an advantageous arrangement of aromatic residues in proteins? Statistical analysis of aromatic interactions in globular proteins. Comput. Struct. Biotechnol. J. 2021; 19: 5960–8. https://doi.org/10.1016/j.csbj.2021.10.036
- Duan S., Wang S., Song Y., Gao N., Meng L., Gai Y., et al. A novel HIV-1 inhibitor that blocks viral replication and rescues APOBEC3s by interrupting vif/CBFβ interaction. J. Biol. Chem. 2020; 295(43): 14592–605. https://doi.org/10.1074/jbc.RA120.013404
- Kardani K., Hashemi A., Bolhassani A. Comparison of HIV-1 Vif and Vpu accessory proteins for delivery of polyepitope constructs harboring Nef, Gp160 and P24 using various cell penetrating peptides. PLoS One. 2019; 14(10): e0223844. https://doi.org/10.1371/journal.pone.0223844
- Delviks-Frankenberry K.A., Ackerman D., Timberlake N.D., Hamscher M., Nikolaitchik O.A., Hu W.S., et al. Development of Lentiviral Vectors for HIV-1 Gene Therapy with Vif-Resistant APOBEC3G. Mol. Ther. Nucleic Acids. 2019; 18: 1023–38. https://doi.org/10.1016/j.omtn.2019.10.024
- Murzakova A., Kireev D., Baryshev P., Lopatukhin A., Serova E., Shemshura A., et al. Molecular epidemiology of HIV-1 subtype G in the Russian Federation. Viruses. 2019; 11(4): 348. https://doi.org/10.3390/v11040348
- Бобкова М.Р. Дефектные провирусы ВИЧ: возможное участие в патогенезе ВИЧ-инфекции. Вопросы вирусологии. 2024; 69(5): 399–414. https://doi.org/10.36233/0507-4088-261 https://elibrary.ru/pselci
- Веселова Е.И., Каминский Г.Д., Самойлова А.Г., Васильева И.А. Резервуар ВИЧ у больных ВИЧ-инфекцией. Туберкулёз и болезни лёгких. 2019; 97(5): 50–7. http://doi.org/10.21292/2075-1230-2019-97-5-50-57 https://elibrary.ru/hfadpt
- Jayaraman B., Fernandes J.D., Yang S., Smith C., Frankel A.D. Highly mutable linker regions regulate HIV-1 rev function and stability. Sci. Rep. 2019; 9(1): 5139. https://doi.org/10.1038/s41598-019-41582-7
- Li L., Dahiya S., Kortagere S., Aiamkitsumrit B., Cunningham D., Pirrone V., et al. Impact of Tat genetic variation on HIV-1 disease. Adv. Virol. 2012; 2012: 123605. https://doi.org/10.1155/2012/123605
- Chen G., He Z., Wang T., Xu R., Yu X.F. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J. Virol. 2009; 83(17): 8674–82. https://doi.org/10.1128/JVI.00653-09
- Williams ME. HIV-1 Vif protein sequence variations in South African people living with HIV and their influence on Vif-APOBEC3G interaction. Eur. J. Clin. Microbiol. Infect. Dis. 2024; 43(2): 325–38. https://doi.org/10.1007/s10096-023-04728-0
- Савченко-Бельский В.Ю., Мальцева М.В., Маслова А.П. Проблемы и перспективы развития транспортной системы Московской агломерации. Транспортное дело России. 2022; (1): 124–7. https://doi.org/10.52375/20728689_2022_1_124 https://elibrary.ru/cctqsp
Дополнительные файлы