Prevalence and phylogenetic analysis of cytomegalovirus (Orthoherpesviridae: Cytomegalovirus: Cytomegalovirus humanbeta5) genetic variants from children and immunocompromised patients in central Russia
- Authors: Van’kova O.E.1, Brusnigina N.F.1, Novikova N.A.1
-
Affiliations:
- Nizhny Novgorod Research Institute of Epidemiology and Microbiology named after Academician I.N. Blokhina
- Issue: Vol 69, No 6 (2024)
- Pages: 535-545
- Section: ORIGINAL RESEARCHES
- URL: https://ogarev-online.ru/0507-4088/article/view/277915
- DOI: https://doi.org/10.36233/0507-4088-277
- EDN: https://elibrary.ru/epnrav
- ID: 277915
Cite item
Full Text
Abstract
Introduction. Cytomegalovirus (CMV) is a DNA-containing virus that is widespread worldwide and is of great importance in infectious pathology of children and adults.
The aim of this study is to evaluate the prevalence of CMV among children and immunocompromised patients in the Nizhny Novgorod region (central Russia) and to perform a phylogenetic analysis of the identified strains.
Materials and methods. DNA samples of CMV detected in frequently ill children and adult recipients of solid organs were studied. The genetic diversity of CMV was assessed for two variable genes: UL55(gB) and UL73(gN), using NGS technology on the Illumina platform. Phylogenetic trees were constructed in the MEGA X program, the reliability of the cluster topology on the trees was confirmed using the rapid bootstrap method based on the generation of 500 pseudo-replicas.
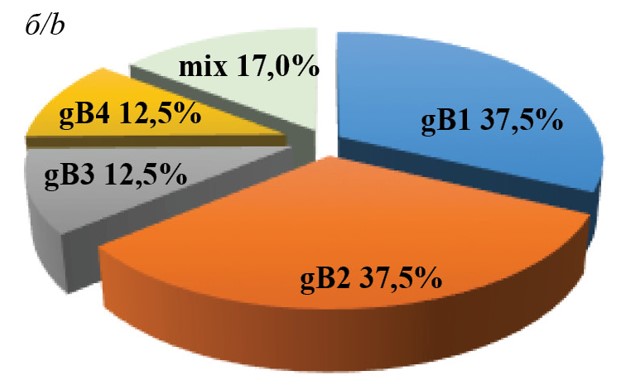
Results. Circulation of 5 CMV genotypes by gene UL55(gB) and 5 genotypes by gene UL73(gN) was established in the territory of the Nizhny Novgorod region. While genotypes gB1 and gB2 dominated both in children and in adults, genotype gB5 was detected only in children. The spectrum of gN genotypes was more diverse: genotypes gN4a and gN3b prevailed in children, and gN1 and gN4b genotypes were predominant in adults. The obtained results allowed us to establish the similarity of the landscape of CMV genotypes circulating in Russia (Nizhny Novgorod region), Brazil, China and the USA.
Conclusion. The obtained data indicate the similarity of the landscape of CMV genotypes circulating in Russia (Nizhny Novgorod region), Brazil, China and the USA: in children, the predominant genotypes are gB1 (40.0%), gB2 (33.3%), gN4a (42.8%), and gN3b (28.6%), while in adults (recipients of solid organs) genotypes gB1 (37.5%), gB2 (37.5%), gN1 (26.3%), and gN4b (26.3%) are prevailed.
Full Text
##article.viewOnOriginalSite##About the authors
Ol’ga E. Van’kova
Nizhny Novgorod Research Institute of Epidemiology and Microbiology named after Academician I.N. Blokhina
Author for correspondence.
Email: voe0@mail.ru
ORCID iD: 0000-0002-9838-1133
art. researcher, laboratory of metagenomics and molecular indication of pathogens
Russian Federation, 603950, Nizhny NovgorodNina F. Brusnigina
Nizhny Novgorod Research Institute of Epidemiology and Microbiology named after Academician I.N. Blokhina
Email: nfbrusnigina@yandex.ru
ORCID iD: 0000-0003-4582-5623
PhD, leading researcher, head of the laboratory of metagenomics and molecular Indication of pathogens
Russian Federation, 603950, Nizhny NovgorodNadezhda A. Novikova
Nizhny Novgorod Research Institute of Epidemiology and Microbiology named after Academician I.N. Blokhina
Email: novikova_na@mail.ru
ORCID iD: 0000-0002-3710-6648
DrSci (Biology), professor, leading researcher, head of the laboratory of molecular epidemiology of viral infections
Russian Federation, 603950, Nizhny NovgorodReferences
- Connoly S.A., Jardetzky T.S., Longnecker R. The structural basis of herpesvirus entry. Nat. Rev. Microbiol. 2021; 19(2): 110–21. https://doi.org/10.1038/s41579-020-00448-w
- Ssentongo P., Hehnly C., Birungi P., Roach M.A., Spady J., Fronterre C., et al. Congenital cytomegalovirus infection burden and epidemiologic risk factors in countries with universal screening. JAMA Netw. Open. 2021; 4(8): e2120736. https://doi.org/10.1001/jamanetworkopen.2021.20736
- Gorbachev V.V., Dmitrachenko T.I., Semenov V.M., Egorov S.K. Aspects of cytomegalovirus reactivation in critically ill patients. Zhurnal infektologii. 2022; 14(4): 61–8. https://doi.org/10.22625/2072-6732-2022-14-4-61-68 https://elibrary.ru/davphd (in Russian)
- Hayden R.T., Caliendo A.M. Persistent challenges of interassay variability in transplant viral load testing. J. Clin. Microbiol. 2020; 58(10): e00782-20. https://doi.org/10.1128/JCM.00782-20
- Mu H., Qiao W., Zou J., Zhang H. Human cytomegalovirus glycoprotein B genotypic distributions and viral load in symptomatic infants. J. Infect. Dev. Ctries. 2023; 17(12): 1806-1813. https://doi.org/10.3855/jidc.18291
- Shakhgildyan V.I. Congenital cytomegalovirus infection: current challenges and possible solutions. Neonatologiya: novosti, mneniya, obuchenie. 2020; 8(4): 61–72. https://doi.org/10.33029/2308-2402-2020-8-4-61-72 https://elibrary.ru/xsnbrk (in Russian)
- Vankova O.E., Brusnigina N.F. Molecular and phylogenetic characteristics of cytomegaloviruses isolated from children in Nizhny Novgorod. Zdorov‘e naseleniya i sreda obitaniya – ZNiSO. 2021; (4): 25–30. https://doi.org/10.35627/2219-5238/2021-337-4-25-30 https://elibrary.ru/rfedkt (in Russian)
- Vankova O.E., Brusnigina N.F. Genotyping clinical cytomegalovirus isolates in solid-organs-transplant recipients. Infektsiya i immunitet. 2022; 12(1): 59–68. https://doi.org/10.15789/2220-7619-GCC-1653 https://elibrary.ru/thvdbq (in Russian)
- Ye L., Qian Y., Yu W., Guo G., Wang H., Xue X. Functional profile of human cytomegalovirus genes and their associated diseases: a review. Front. Microbiol. 2020; 11: 2104. https://doi.org/10.3389/fmicb.2020.02104
- Marti-Carreras J., Maes P. Human cytomegalovirus genomics and transcriptomics through the lens of next-generation sequencing: revision and future challenges. Virus Genes. 2019; 55(2): 138–64. https://doi.org/10.1007/s11262-018-1627-3
- Sijmons S., Thys K., Ngwese M.M., Damme V.E., Dvorak J., Loock M.V., et al. High-throughput analysis of human cytomegalovirus genome diversity highlights the widespread occurrence of gene-disrupting mutations and pervasive recombination. J. Virol. 2015; 89(15): 7673–95. https://doi.org/10.1128/jvi.00578-15
- Dong N., Cao L., Zheng D., Su L., Lu L., Dong Z., et al. Distribution of CMV envelop glycoprotein B, H and N genotypes in infants with congenital cytomegalovirus symptomatic infection. Front. Pediatr. 2023; 11: 1112645. https://doi.org/10.3389/fped.2023.1112645
- Vankova O.E., Brusnigina N.F., Novikova N.A. NGS technology in monitoring the genetic diversity of cytomegalovirus strains. Sovremennye tekhnologii v meditsine. 2023; 15(2): 41–7. https://doi.org/10.17691/stm2023.15.2.04 https://elibrary.ru/gwepec (in Russian)
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018; 35(6): 1547–9. https://doi.org/10.1093/molbev/msy096
- Sarkar A., Das D., Ansari S., Chatterjee R. P., Mishra L., Basu B., et al. Genotypes of glycoprotein B gene among the Indian symptomatic neonates with congenital CMV infection. BMC Pediatr. 2019; 19(1): 291. https://doi.org/10.1186/s12887-019-1666-5
- Puhakka L., Pati S., Lappalainen M., Lonnqvist T., Niemensivu R., Lindahl P., et al. Viral shedding and distribution of cytomegalovirus glycoprotein H(UL75), glycoprotein B (UL55), and glycoprotein N(UL73) genotypes in congenital cytomegalovirus infection. J. Clin. Virol. 2020; 125: 104287. https://doi.org/10.1016/j.jcv.2020.104287
- Pati S., Pinninti S., Novak Z., Chowdhury N., Patro R., Fowler K., et al. Genotypic diversity and mixed infection in newborn disease and hearing loss in congenital cytomegalovirus infection. J. Pediatr. Infect. Dis. 2014; 32(10): 1050–4. https://doi.org/10.1097/inf.0b013e31829bb0b9
- Wu X.J., Wang Y., Zhu Z.L., Xu Y., He G.S., Han Y., et al. The correlation of cytomegalovirus gB genotype with viral DNA load and treatment time in patients with CMV infection after hematopoietic stem cell transplantation. Zhonghua Xue Ye Xue Za Zhi. 2013; 34(2): 109–12. (in Chinese)
- Pignatelli S., Lazzarotto Т., Gatto M.R. Cytomegalovirus gN genotypes distribution among congenitally infected newborns and their relationship with symptoms at birth and sequelae. Clin. Infect. Dis. 2010; 51(1): 33–41. https://doi.org/10.1086/653423
- Jankovića M., Ćupića M., Kneževića A., Vujićb D., Soldatovićc I., Zečevićb Ž., et al. Cytomegalovirus glycoprotein B and N genotypes in pediatric recipients of the hematopoietic stem cell transplant. J. Virology. 2020; 548: 168–73. https://doi.org/10.1016/j.virol.2020.07.010
- Wang H., Valencia S.M., Preifer S.P., Jensen J.D., Kowalik T.F., Permar S.R. Common polymorphisms in the glycoproteins of human cytomegalovirus and associated strain-specific immunity. Viruses. 2021; 13(6): 1106. https://doi.org/10.3390/v13061106
- Suárez N.M., Wilkie G.S., Hage E., Camiolo S., Holton M., Hughes J., et al. Human cytomegalovirus genomes sequenced directly from clinical material: variation, multiple-strain infection, recombination, and gene loss. J. Infect. Dis. 2019; 220(5): 781–91. https://doi.org/10.1093/infdis/jiz208
Supplementary files